Abstract
Introduction: Conventional karyotype analysis is one of the most important diagnostic tools to determine the prognosis of acute myeloid leukemia (AML), in which more than 50% of cases are affected. However, the low sensitivity of this technique hampers the detection of small genetic alterations like Copy Number Variation (CNV) that could affect the pathophysiology and prognosis of the disease. Current modern genomic technologies based on next generation sequencing (NGS) are capable to detect CNV at low frequencies.
Objective: To analyse CNV of genes related to myeloid neoplasms profile in AML patients at diagnosis and evaluate their connection with the mutational profile, and its possible influence on the clinical-biological phenotype and prognosis of the disease.
Materials and methods: The CNV and mutational profile were analysed in samples from 380 AML patients, from PLATAFO-LMA reference centres (IMIBIC, Córdoba and Dr Negrín Las Palmas de Gran Canaria) by NGS, applying a panel of 30 genes (154 regions) related to myeloid neoplasms (Sophia Myeloid Solution®) on Ilumina Myseq platform.
Results: NGS detected CNV in at least one gene in 103 AML patients (27.1%). NGS detected 103 gains and 206 losses of genetic material. The median number of genes affected by CNV was 2 (range 1-12). When comparing with conventional karyotype information, CNV provided additional information in 51% of the cases.
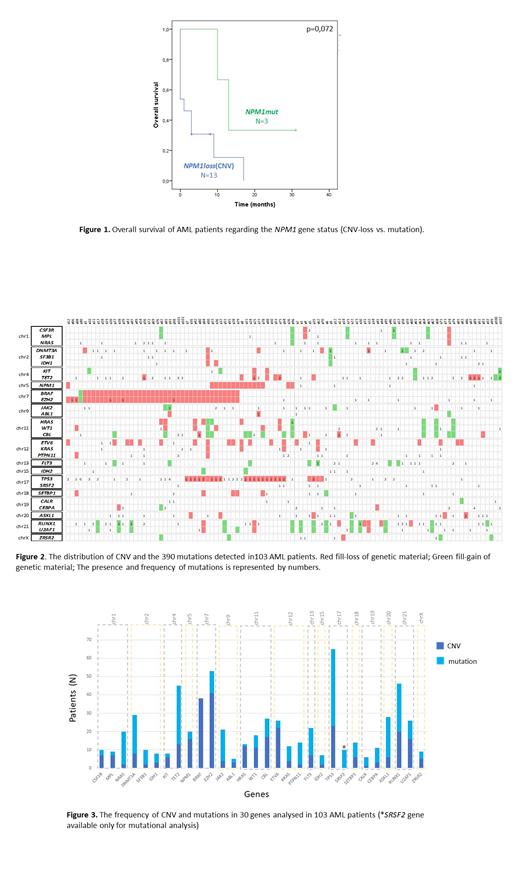
The chromosomes 7, 11 and 21 were most affected with CNV, occurring in 79 (76.7%), 40 (38.8%) and 36 (35%) patients, respectively. The gains of genetic material occurred more frequently on chromosome 21 in U2AF1 and RUNX1 genes, in 13 patients each. The loss of genetic material in EZH2 and BRAF genes occurred mutually. Interestingly, we observed the tendency that patients with CNV (loss) in NPM1 gene had shorter overall survival compared to cases with NPM1 mutated and without CNV in this gene (1 month vs. 13 months, p = 0.072) (Figure 1). Moreover CNV (loss) in TP53 gene was associated with mutations in this gene, other than deletions (p <0.05).
In addition, NGS detected 390 mutations distributed in 29 genes in 103 AML with CNV. The median number of mutations was 3 (range 1-10) (Figure 2). Furthermore, only 2 patients did not have any mutation in genes analysed. The distribution and frequency of genes affected by CNV and by mutations was different (Figure 3).
Conclusions: The CNV of genes related to myeloid neoplasms are frequent in AML patients (27.1%) and provides additional information to the conventional karyotype in half of the cases. The loss of NPM1 gene could affect survival of AML patients. The use of NGS with CNV analysis provides important information on copy number alterations that are not detected by the karyotype, which could significantly affect the pathophysiology of AML and with potential clinical impact, especially in patients with normal karyotype.
Hernández Rivas: Pfizer: Honoraria, Membership on an entity's Board of Directors or advisory committees; Novartis: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Celgene/BMS: Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Amgen: Membership on an entity's Board of Directors or advisory committees.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal